Imagine the ability to search through hundreds of millions of high-dimensional vectors in a split second, all while using less storage and memory than ever before. This might sound impossible, but with binary vectors—–OpenSearch’s latest advancement in large-scale vector search—–it’s a reality. In a world where data is growing at explosive rates, handling massive datasets with reduced memory is crucial, whether you’re building recommendation systems or advanced search engines. In this blog post, we’ll explore how binary vectors perform as compared to traditional FP32 vectors, especially with large datasets like our randomly generated 768-dimensional, 100-million vector dataset. We’ll look at storage, memory usage, and search speed as well as how binary vectors might change the way you approach vector search.
What’s the difference between FP32 and binary vectors?
FP32 vectors have long been the standard for vector search because of their high precision and seamless integration with many large language models (LLMs), which typically produce vectors in floating-point formats. However, this precision comes at a price—increased storage and memory. As your data needs increase, this trade-off can be hard to justify. In contrast, binary vectors use only 1s and 0s, as shown in the following image.

This binary format makes binary vectors more compact and faster to process. LLMs are increasingly generating binary embeddings in order to improve their efficiency on large datasets, offering significant reductions in storage, memory, and latency.
Using binary vectors in OpenSearch
Let’s explore how you can use binary vectors in your OpenSearch solution.
Data preparation
To get started, you’ll need binary vector data. Luckily, many models now generate embeddings in binary format. For example, the Cohere Embed v3 model generates binary vectors.
Binary vectors are arrays of 1s and 0s, such as [0, 1, 1, 0, 1, 1, 0, 0, 1, 0, 0, 0, 1, 1, 0, 0]. However, OpenSearch requires that binary vectors be packed into an int8 byte format. For example, the preceding bit array would be converted to [108, -116], as shown in the following image.
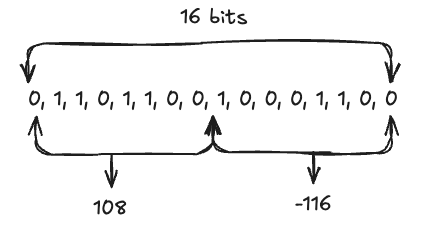
Many embedding models already generate binary vectors in int8 byte format, so extra packing is usually unnecessary. However, if your data is stored in a bit array, you can easily convert it into a byte array using the numpy library:
import numpy as np
bit_array = [0, 1, 1, 0, 1, 1, 0, 0, 1, 0, 0, 0, 1, 1, 0, 0]
bit_array_np = np.array(bit_array, dtype=np.uint8)
byte_array = np.packbits(bit_array_np).astype(np.int8).tolist()
Ingestion and search
Once your data is stored in byte arrays, you’ll need to ingest those into OpenSearch.
First, set the data type to binary in your index mapping and ensure that the vector dimensions are a multiple of 8 (if they are not, pad the vectors with zeros):
PUT /test-binary-hnsw
{
"settings": {
"index": {
"knn": true
}
},
"mappings": {
"properties": {
"my_vector": {
"type": "knn_vector",
"dimension": 16,
"data_type": "binary",
"space_type": "hamming",
"method": {
"name": "hnsw",
"engine": "faiss"
}
}
}
}
}
Binary vectors in OpenSearch use the Hamming distance for indexing and search.
Next, pack the binary vector into a byte format for both indexing and searching. Otherwise, using binary vectors is similar to working with FP32 vectors. In the following example, you’ll index two documents with vector values of [0, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 0, 1, 0, 0, 0] and [0, 0, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 1, 0, 1, 1]:
PUT _bulk
{"index": {"_index": "test-binary-hnsw", "_id": "1"}}
{"my_vector": [7, 8]}
{"index": {"_index": "test-binary-hnsw", "_id": "2"}}
{"my_vector": [10, 11]}
Finally, search for the vector closest to a query vector of [0, 1, 1, 0, 1, 1, 0, 0, 1, 0, 0, 0, 1, 1, 0, 0]:
GET /test-binary-hnsw/_search
{
"size": 1,
"query": {
"knn": {
"my_vector": {
"vector": [108, -116],
"k": 1
}
}
}
}
Performance comparison
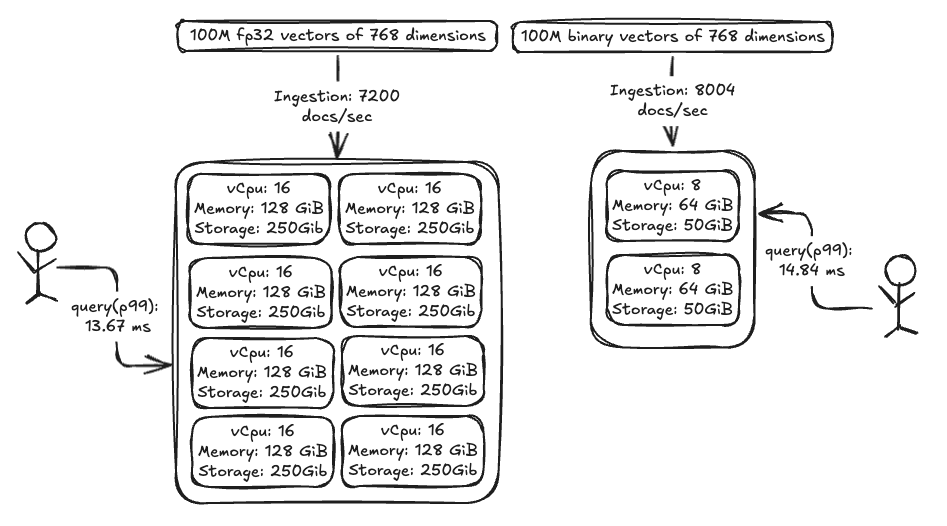
Now let’s look at the resource savings realized by using binary vectors. In our benchmarking tests, we observed similar ingestion speeds and query times between FP32 and binary vectors, even while using 8x less powerful hardware for binary vectors, as shown in the following image.
Cluster setup
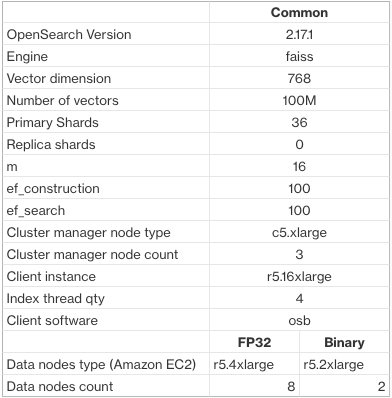
Our benchmark used a randomly generated 100-million vector dataset with 768 dimensions, comparing FP32 and binary vectors. The clusters were identical except for the data nodes: binary vectors used nodes that were 2x smaller and 4x fewer, leading to an 86% cost reduction. The following table outlines the benchmarking setup.
Performance results
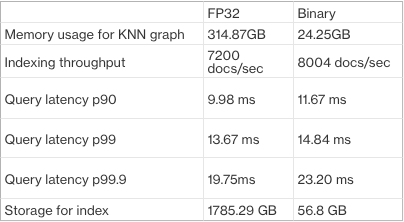
Even on 8x smaller hardware, binary vectors delivered indexing speeds and query times comparable to FP32 vectors on more powerful machines. With binary vectors, memory usage was reduced by 92% and storage by 97%, producing significant savings. The results are presented in the following table.
Accuracy
In terms of recall, you can expect around 0.97 recall compared to exact search. OpenSearch uses the HNSW algorithm for approximate nearest neighbor searches, but the accuracy of the results depends on your dataset. Some models produce binary vectors with high accuracy: for example, Cohere Embed v3 reports a 94.7% match in search quality compared to FP32 embeddings. Thus, when using a model producing quality binary embeddings, binary vectors can reach nearly the same accuracy as FP32 vectors.
Binary vector challenges: When they fall short
When your model only produces FP32 vectors but you’d like to use binary vectors in OpenSearch, the process can get a little complicated. The following example demonstrates using binary vector search in OpenSearch with FP32 vectors, along with the challenges involved.
For this example, we used the Cohere Simple dataset from Hugging Face. Because the data was in FP32 format, we converted it to binary format by setting zero and negative values to 0 and positive values to 1. The following image illustrates the conversion process.
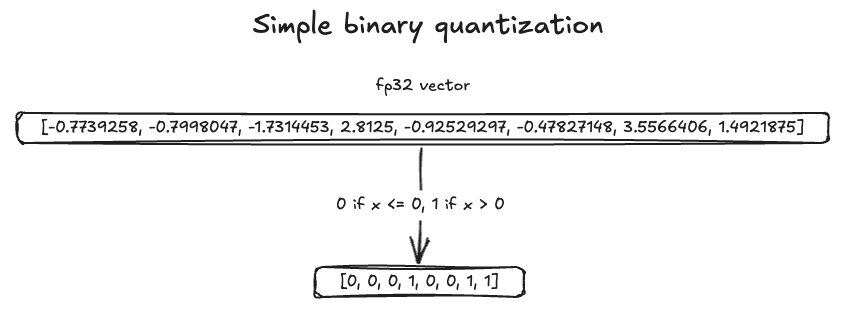
In terms of recall, binary vectors achieved a score of 0.73196. To reach a recall of 0.93865, we needed 3x oversampling. Additionally, rescoring requires storing the original vector format, which adds disk usage unless the original vector is stored outside of OpenSearch. The following image illustrates oversampling with rescoring. Note that recall may vary across datasets.
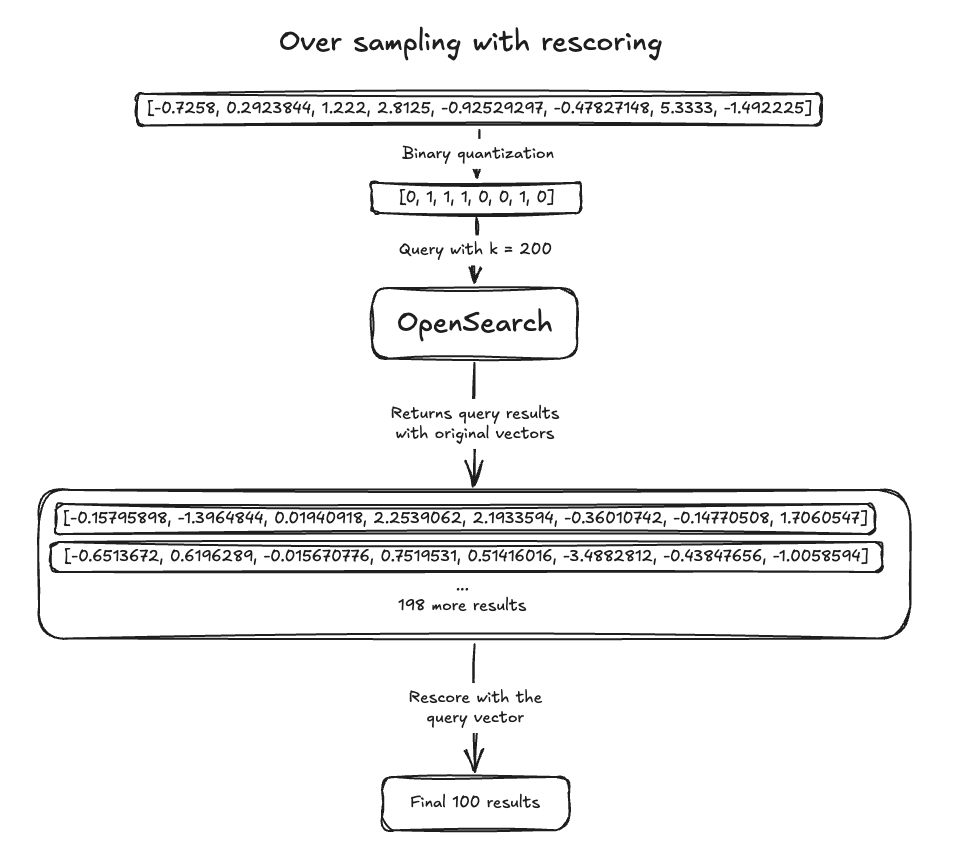
By quantizing, oversampling, and rescoring, binary vectors can achieve similar recall to FP32 vectors while using significantly less memory. However, managing these steps outside of OpenSearch can be cumbersome. Disk-based vector search simplifies the process by automatically using advanced quantization techniques for all required steps. Give it a try and see the difference!
Conclusion
Binary vectors offer an efficient alternative to FP32 vectors, reducing memory and storage usage by more than 90% while maintaining strong performance on smaller hardware. This efficiency makes binary vectors ideal for large-scale vector search applications like recommendation systems or search engines, where speed and resource savings are critical. If you’re handling massive datasets, binary vectors provide a practical way to scale search capabilities without increasing costs.
What’s next?
Binary vector support is now available in OpenSearch 2.16 and later. Check out the OpenSearch binary vector documentation for detailed instructions.
If you’re working with floating-point vectors, disk-based vector search provides the memory efficiency of binary vector search without losing recall. It performs binary quantization, oversampling, and rescoring automatically, all while maintaining the low memory usage of binary vector search.
We encourage you to try binary vectors for yourself and to stay tuned for our upcoming blog post on disk-based vector search.